
What if generative AI could design life-saving antibiotics, not just art and text? In a new Cell Biomaterials paper, Penn researchers introduce AMP-Diffusion, a generative AI tool used to create tens of thousands of new antimicrobial peptides (AMPs) — short strings of amino acids, the building blocks of proteins — with bacteria-killing potential. In animal models, the most potent AMPs performed as well as FDA-approved drugs, without detectable adverse effects.
While past breakthroughs at Penn have shown that AI can successfully sort through mountains of data to identify promising antibiotic candidates, this study adds to a small but growing number of demonstrations that AI can invent antibiotic candidates from scratch.
“Nature’s dataset is finite; with AI, we can design antibiotics evolution never tried,” says César de la Fuente, Presidential Associate Professor in Bioengineering (BE) and in Chemical and Biomolecular Engineering in the University of Pennsylvania School of Engineering and Applied Science (Penn Engineering), in Psychiatry and Microbiology in the Perelman School of Medicine and in Chemistry in the School of Arts & Sciences, and the paper’s senior co-author.
“We’re leveraging the same AI algorithms that generate images, but augmenting them to design potent new molecules,” adds Pranam Chatterjee, Assistant Professor in BE and in Computer and Information Science within Penn Engineering, and the paper’s other senior co-author, who began work on the project while at Duke University.
Two Labs, One Goal
For years, de la Fuente’s lab has successfully leveraged AI to search for molecules with antimicrobial properties in unlikely places, from the proteins of woolly mammoths to those of animal venom and ancient microbes called archaea. “Unfortunately, antibiotic resistance keeps increasing faster than we can discover new antibiotic candidates,” says de la Fuente.
That led to his lab teaming up with Chatterjee’s, which typically designs peptides using AI to treat diseases for which conventional methods of drug development have fallen short. “It seemed like a natural fit,” says Chatterjee. “Our lab knows how to design new molecules using AI, and the de la Fuente Lab knows how to identify strong antibiotic candidates using AI.”
Tuning Out the Noise
While some generative AI models, like ChatGPT, work by predicting the next word or element in a sequence, “diffusion” models start from random “noise” and iteratively refine it into a coherent output — the principle behind tools like DALL·E and Stable Diffusion.
AMP-Diffusion works the same way, only instead of “denoising” pixels, it refines sequences of amino acids. “It’s almost like adjusting the radio,” says de la Fuente. “You start with static, and then eventually the melody emerges.”
At least two other research teams have applied diffusion models to design antimicrobial peptides, but AMP-Diffusion takes a novel approach.
Instead of first training its own protein “latent space” — a kind of internal map of how proteins are structured — AMP-Diffusion builds on ESM-2, a widely used protein language model from Meta trained on hundreds of millions of natural protein sequences.
Because ESM-2 already has a rich “mental map” of how real proteins fit together, AMP-Diffusion doesn’t need to relearn basic biology. That means it can generate candidate AMPs faster, and its outputs are more likely to follow the intricate patterns that make peptides effective.
Chatterjee’s team also designed AMP-Diffusion to consult ESM-2’s built-in rules while “denoising,” essentially giving the new tool a coach that keeps it grounded in biological reality.
“Instead of teaching the model the ABCs of biology, we started with a fluent speaker,” says Chatterjee. “That shortcut lets us focus on designing peptides with a real shot at becoming drugs.”
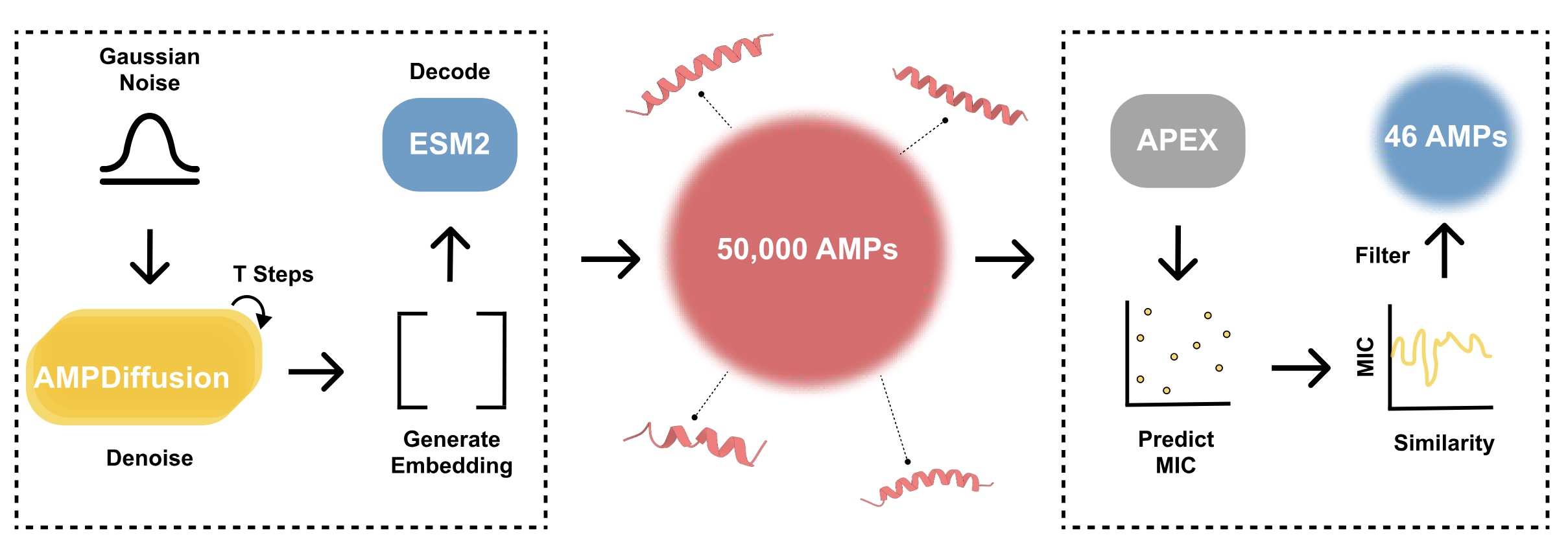
From 50,000 Designs to Two in vivo Winners
Using AMP-Diffusion, the researchers generated the amino-acid sequences for about 50,000 candidates. “That’s far more candidate drugs than we could ever test,” says de la Fuente. “So we used AI to filter the results.”
Fine-tuned by hunting for antibiotic candidates everywhere from the proteins of ancient microbes to those of Neanderthals, APEX 1.1, an AI tool developed by de la Fuente’s lab, ranked the candidate AMPs according to a number of criteria. These included predicting which sequences would have strong bacteria-killing power, filtering out peptides that were too similar to known AMPs and ensuring the remaining candidates covered a diverse range of sequence types.
After synthesizing the 46 most promising candidates, the de la Fuente lab tested them in human cells and animal models. Treating skin infections in mice, two AMPs demonstrated efficacy on par with levofloxacin and polymyxin B, FDA-approved drugs used to treat antibiotic-resistant bacteria, without adverse effects. “It’s exciting to see that our AI-generated molecules actually worked,” says Chatterjee. “This shows that generative AI can help combat antibiotic resistance.”
Next Steps for AI-Generated Antibiotics
In the future, the researchers hope to refine AMP-Diffusion, giving it the capability to denoise with a more specific goal in mind, like treating a particular type of bacterial infection, among other features. “We’ve shown the model works, and now if we can steer it to enhance beneficial drug-like properties, we can make ready-to-go therapeutics,” says Chatterjee.
For the researchers, the current study is a proof of principle: generative AI can move beyond mining what evolution has already created to actually designing new antibiotics. “Ultimately, our goal is to compress the antibiotic discovery timeline from years to days,” says de la Fuente.
Pranam Chatterjee acknowledges funding from the Hartwell Individual Biomedical Award, as well as NIH grants R35GM155282 and R21-CA278468. He is a co-founder of Gameto, Inc., UbiquiTx, Inc., and AtomBioworks, Inc., and serves as an advisor to various peptide therapeutics companies.
César de la Fuente-Nunez holds a Presidential Professorship at the University of Pennsylvania and acknowledges funding from the Procter & Gamble Company, United Therapeutics, a BBRF Young Investigator Grant, the Nemirovsky Prize, Penn Health-Tech Accelerator Award, Defense Threat Reduction Agency grants HDTRA11810041 and HDTRA1-23-1-0001, and the Dean’s Innovation Fund from the Perelman School of Medicine at the University of Pennsylvania. Research reported in this publication was supported by the Langer Prize (AIChE Foundation), the NIH R35GM138201, and DTRA HDTRA1-21-1-0014.
Additional co-authors include first author Marcelo D.T. Torres of Penn Medicine, Tianlai Chen of Duke, and Fangping Wan of Penn Engineering.
